Server manual
Selection of postsynaptic (PS) proteins
We downloaded the human reference proteome from UniProt then used four resources to define the postsynaptic proteome: SynaptomeDB, G2C, SynGO and Gene ontology annotations. For all proteins listed in any of these resources we collected interactions from various sources.
Furthermore, we also display protein expression data from Human Protein Atlas, however we are not using it as a filter. Proteins showing neuronal expression in Cerebral cortex, Caudate or in the Hippocampus are displayed.
A comprehensive collection of PS protein-protein interactions (PPIs)
We collected nearly 2000 experiments from the literature describing PPIs using at MIMIx or higher standards. During the collection we aimed to look for papers that were not included in other similar resources. To have a complete collection of PS Interactome we also display interactions from IntAct IntAct and BIOGRID databases. In addition, we scanned the PDB database for complexes that have interacting residues. Furthermore, computational evidence from the STRING database is also displayed (above 0.8 cutoff score).
Users can also search for the interaction of any protein. The result page will help the user select proteins that localize to PS.
PSINDB displays the interaction between human PS proteins. We considered evidence from mouse and rat experiments too – in this case the interaction is always mapped back the orthologous human protein, using OMA orthologous database and gene names.
Navigating on the PSINDB database
Data on the PSINDB database can be viewed using three distinct layouts, each displaying more specific information about the PS Interactome. The first level is the search/browse page, where proteins of preconstructed sets can be viewed (or any other search can be performed). The next level is the protein page, where general information is displayed about the protein, along with a summary of their interacting partners. Finally, the interaction page displays every specific experimental (or computational) information about the selected interaction.
Search/browse page
Users can search for proteins using UniProt identifier, UniProt AC, GeneID, Gene name, Ensemble Protein identifier and HGNC codes using the search bar at the top of the page. After searching the results will be listed, along with the number of interactions and the evidence that the protein of choice localizes into the PS.
Users can also search for the interaction of any protein. The result page will help the user select proteins that localize to PS.
We also prepared sets of proteins, e.g, proteins are known to be going through phase separation, proteins with PPIs having literature evidence or transmembrane proteins and more. These sets can be accessed through the browse menu.
Entry page
Function
For each protein basic information about the protein is displayed using UniProt functional annotations.
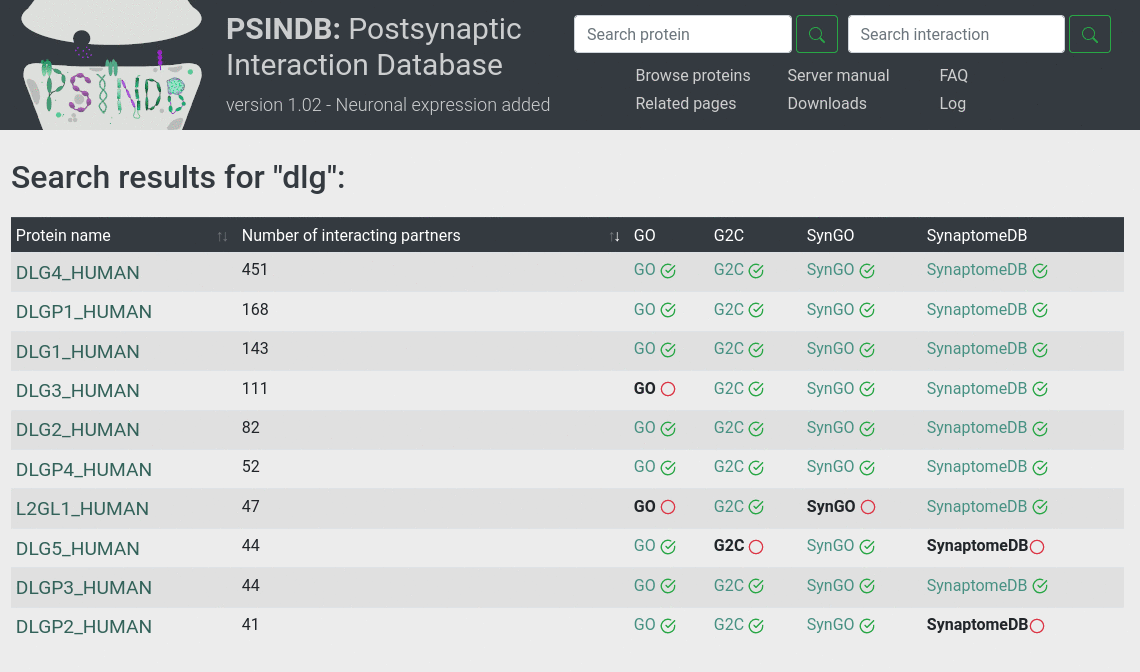
Protein features
Protein features display structural and functional information about the protein mirrored to the protein sequence. The legend below the Features guides the user on how to interpret this information. Using the scroll users can zoom into regions. Clicking into a region, the segment is highlighted along with all other features.
Topology: transmembrane segments were predicted using CCTOP. Since CCTOP also includes experimental evidence, the corresponding entry is linked to the Human Transmembrane Proteome database.
Phase separation: displays protein regions going through phase separation from PhaSePro database. Although currently the number of annotated proteins is low, we expect a rapid increase of annotated data as the field emerges.
Linear motifs: short linear motifs are compact protein regions usually between 3-10 residues playing crucial roles in cell regulation. The ELM database is the most comprehensive collection of experimentally verified motifs. More information about these segments can be found below at the Linear motif section of the entry page.
Phosphorylation: phosphorylations are post-translational modifications that can have a diverse range of functions. They often regulate PPIs by working as switches, changing between the non-modified (OFF) state and the modified (charged) (ON) state. Phosphorylation annotation were collected from UniProt.
PFAM: PFAM is a collection and classification of protein families and domains.
Coiled coil: Coiled-coils are oligomeric helical structural units in proteins connected to a wide range of functions. They can bridge large distances and connect proteins at different sides of large supramolecular structures. Coiled coil segments were predicted using DeepCoil.
Anchor: Intrinsically disordered regions often bind to ordered globular domains. Such disordered binding segments are predicted using Anchor.
Disordered regions: Intrinsically disordered regions (IDRs) are flexible protein regions that can serve various functions. IDRs can mediate binary interactions of proteins or promote their organization of large assemblies. Disordered segments were predicted using IUPred.
Interacting regions: In this panel we mirrored the interaction regions responsible for binding the protein’s partners. Experiments provide different levels of information about the interacting regions. If two or more experiments overlap, we always display the information from the most specific one.
Binding-associated region (source: Intact, PSINDB): a region of a molecule or a component of a complex identified as being involved in an interaction. This may or may not be a region of the molecule in direct contact with the interacting partner.
Sufficient binding region (Intact, PSINDB, PDB): binding will occur when this sequence range is present within a molecule or part of a molecule. This region will contain the direct binding region but may be longer.
Necessary binding region (Intact, PSINDB): a sequence range within a molecule identified as being absolutely required for an interaction. The sequence may or may not be in direct physical contact with the interaction partner.
Atomic contact (PDB): atomic contact between residues. After reconstructing the full PDB structures using the biomatrix information we used Voronota to detect residues establishing interchain interactions.
For experiments on orthologous proteins even if the binding segment was defined, we did not map it back to the original sequence – sometimes this task is trivial, however there are several cases when the alignment is ambiguous, especially when there are disordered segments involved.
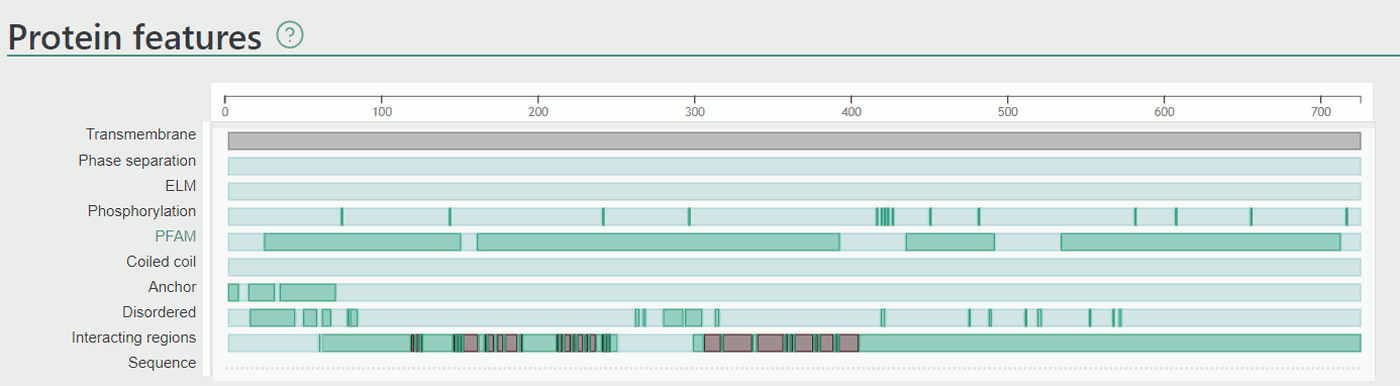
Partners with known interacting regions
This section displays all partners where the interacting region of at least one of the partners was narrowed from the full length sequence. This annotation has the same levels as described earlier in the Protein features/Interacting regions section. The link of the partner navigates to the interaction page with detailed information about the experiments.
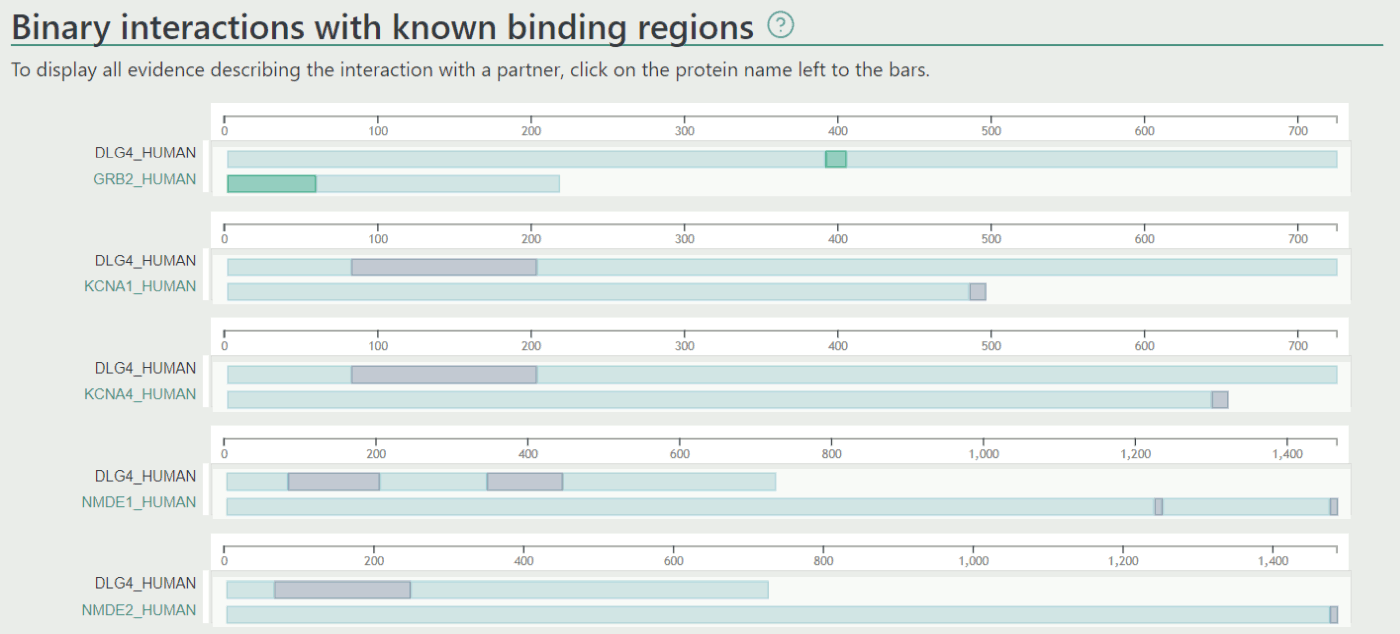
Network
The Network view displays interaction partners and also shows how these partners interact with each other. The PS exhibits an extremely high number of PPIs, therefore we reduced the number of displayed partners to 50 in this view (users can still download the full network from the link below the panel). For this purpose we scored each interaction using the following method: for each evidence we assigned a score. The score of any interaction is the sum of individual pieces of evidence.
| Experiment on human proteins | Experiment on orthologous proteins | |
|---|---|---|
| Low throughput | 1 | 0.8 |
| High throughput | 0.8 | 0.64 |
| Computational | 0.6 | x |
For the definition of low throughput, high throughput and computational evidence see the "All partners" section.
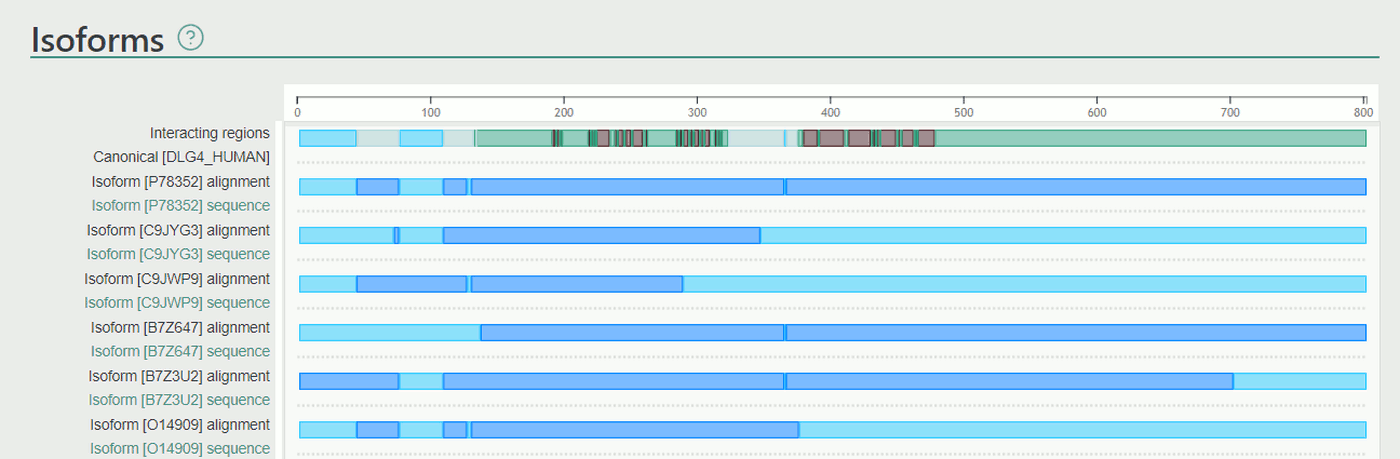
Isoforms
This section displays all isoforms, together with the interacting regions of the canonical form, thus isoforms with missing binding regions can be easily captured. Isoforms were collected from UniProt and they were aligned using ClustalOmega.
Disease-causing germline mutations
Disease-causing germline mutations were collected from the OMIM database. We also highlight mutations that fall into the interacting region of the protein and display partners binding through that region.
Linear motifs
Short linear motifs are compact protein regions usually between 3-10 residues playing crucial roles in cell regulation. The ELM database is the most comprehensive collection of experimentally verified motifs. We also highlight SLiMs that fall into the interacting region of the protein and display partners binding through that region.

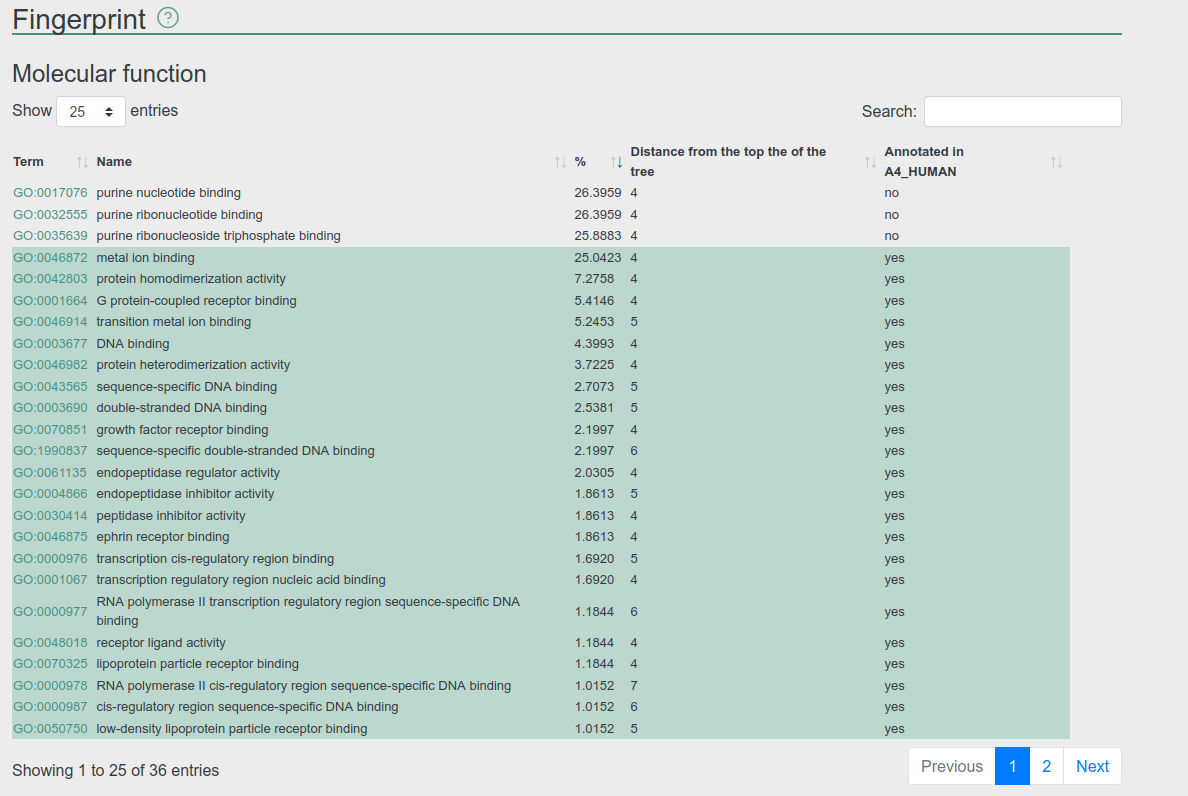
Fingerprint
Fingerprint provides a collection of ontology terms re-occurring in the protein or its interaction partners. We collected all terms annotated for the proteins and also mapped ancestor terms for the given protein. The annotation of different proteins may vary and thus any statistics would be biased, therefore we only display the percentage of different terms in the network and do not calculate further statistics. We also highlight terms that are associated with the entry.
Hits can be filtered using how specific is the given term. Currently PSINDB displays three ontologies: GO: biological process, GO: molecular function and DOID: disease ontology.
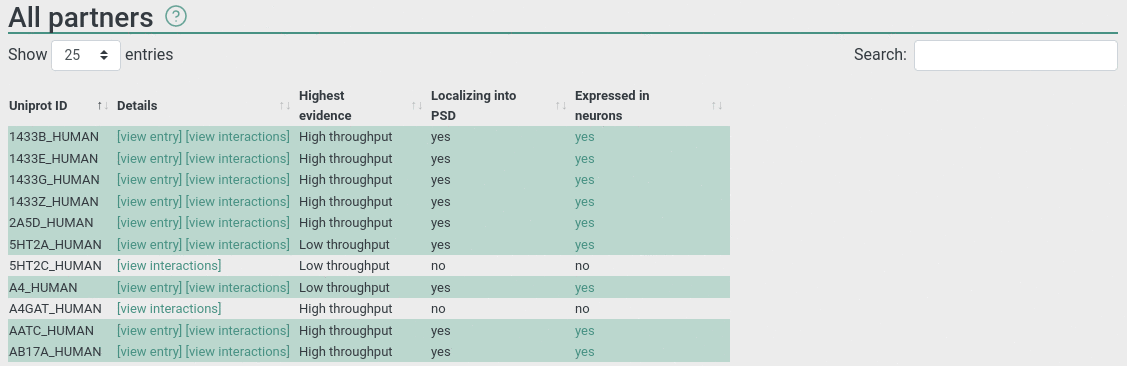
All partners
This panel displays all interacting partners, including the ones that not localize to PS according to our current knowledge. PS partners are highlighted.
For each partner there are two links: [view entry] navigate to the entry page of the selected protein. [view interaction] navigates to the interaction page with detailed information about the experiments.
Highest evidence display the highest level of information about the particular interaction:
*Low throughput (LTP): in case of PSINDB, the curator can make the call to classify the given interaction. In the case of Intact and BIOGRID, we followed the following procedure: in any paper, if using the same experimental setup less than ten experimental data was produced, we assigned it as low throughput. Interactions from PDB are always classified as low throughput.
*High throughput (HTP): in case of PSINDB, the curator can make the call to classify the given interaction. In the case of Intact and BIOGRID, we followed the following procedure: in any paper, if using the same experimental setup ten or more experimental data was produced, we assigned it as high throughput.
Computational: Interactions collected from the STRING database belong to this category.
*We are aware that this classification may have many pitfalls, however this seemed the most efficient way to filter out primary screens. In several cases these interactions are further validated in the same paper, therefore they will get the LTP tag.
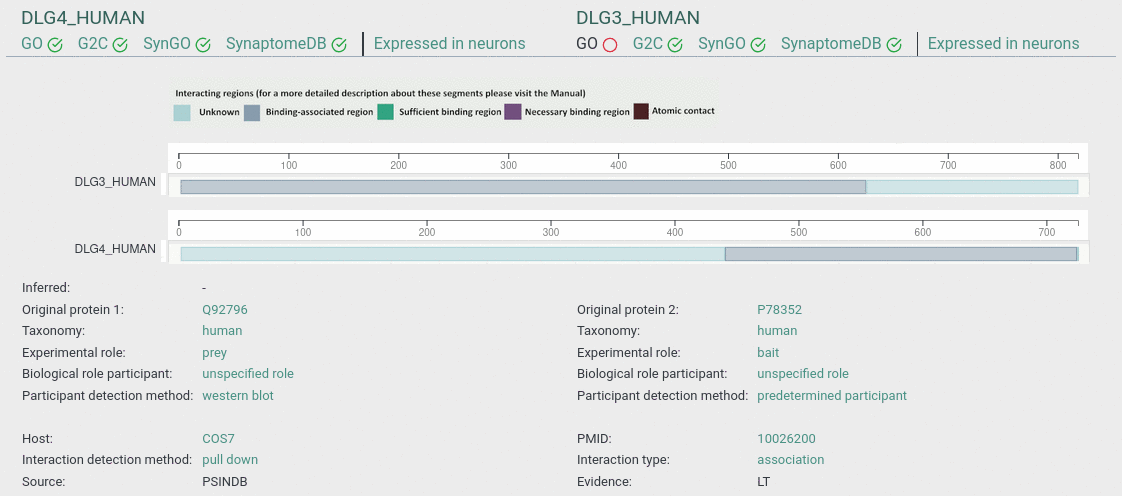
Interaction page
This page contains all evidence of interaction for a pair of proteins. Information is stored according to the PSI MI controlled vocabularies and can be inspected on the Ontology Lookup Services.
Download
Data from PSINDB can be downloaded in the following formats:
- experimental_mitab25.tsv - MITAB 2.5 format file for all experimental data
- PSINDB_mitab25.tsv - MITAB 2.5 format file containing data collected by our curation team
- merged_binding.txt - this file contains the interacting regions, whenever it is available. For interactions with multiple experiments and different regions the union of the interacting region is shown
- postsynaptic_proteins.tsv - List of postsynaptic proteins
- postsynaptic_network.tsv - Full PS network with scores